Think Summer: Introduction — 2023
Submission
Students need to submit the following file by 10:00PM EST through Gradescope inside Brightspace.
-
A Jupyter notebook (a
.ipynbfile).
We’ve provided you with a template notebook for you to use. Please carefully read this section to get started.
|
When you are finished with the project, please make sure to run every cell in the notebook prior to submitting. To do this click . Next, to export your notebook (your |
Project
Motivation: SQL is an incredibly powerful tool that allows you to process and filter massive amounts of data — amounts of data where tools like spreadsheets start to fail. You can perform SQL queries directly within the R environment, and doing so allows you to quickly perform ad-hoc analyses.
Context: This project is specially designed for Purdue University’s Think Summer program, and is coordinated by The Data Mine.
Scope: SQL, SQL in R
Dataset
The following questions will use the imdb database found in Anvil, our computing cluster.
This database has 6 tables, namely:
akas, crew, episodes, people, ratings, and titles.
You have a variety of options to connect with, and run queries on our database:
-
Run SQL queries directly within a Jupyter Lab cell.
-
Connect to and run queries from within R in a Jupyter Lab cell.
-
From a terminal in Anvil.
For consistency and simplicity, we will only cover how to do (1) and (2).
First, for both (1) and (2) you must launch a new Jupyter Lab instance. To do so, please follow the instructions below.
-
Open a browser and navigate to ondemand.anvil.rcac.purdue.edu, and login using your ACCESS credentials. You should be presented with a screen similar to figure (1).
 Figure 1. OnDemand
Figure 1. OnDemand -
Click on "My Interactive Sessions", and you should be presented with a screen similar to figure (2).
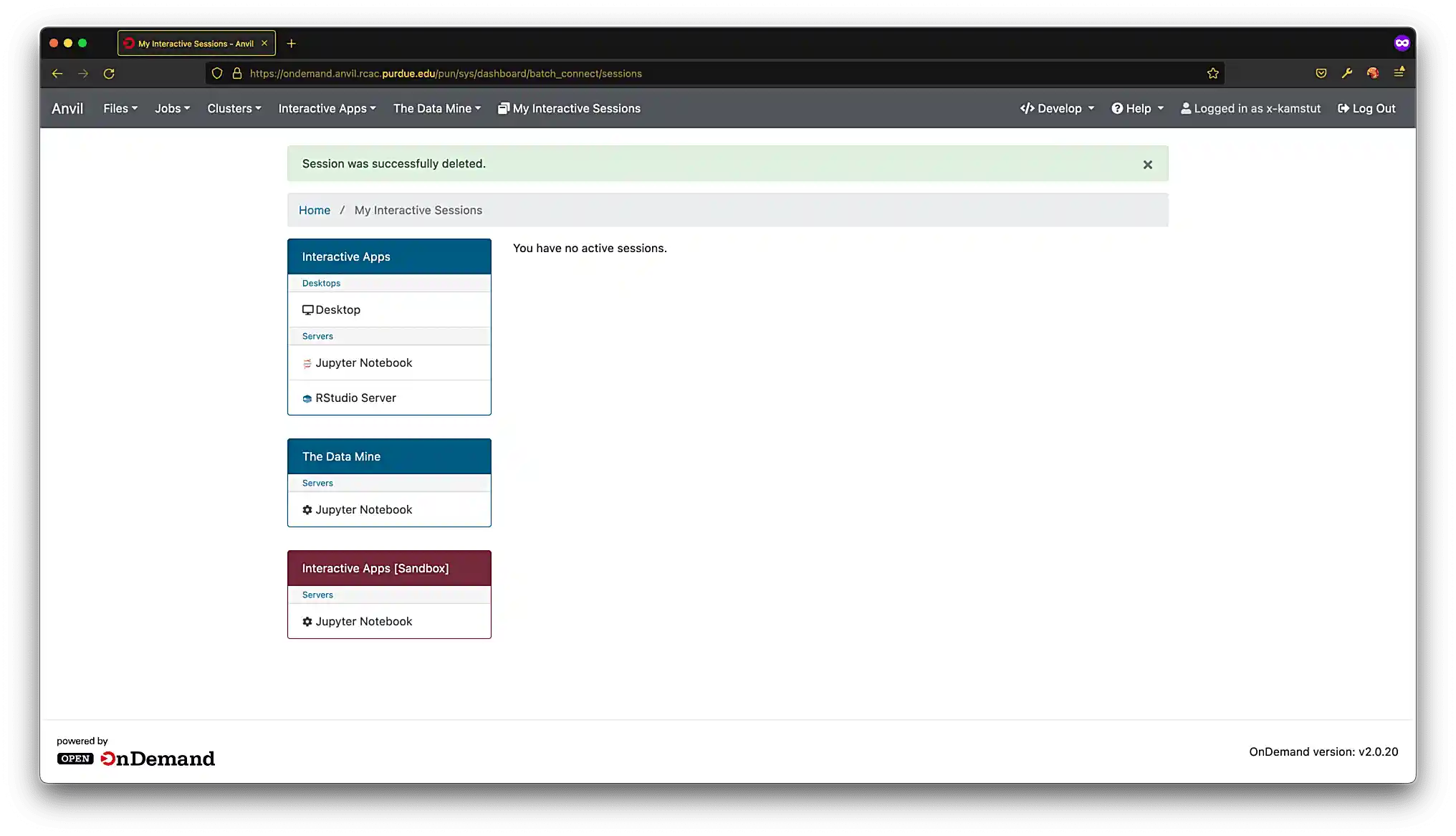 Figure 2. Your interactive Anvil sessions
Figure 2. Your interactive Anvil sessions -
Click on Jupyter Notebook in the left-hand menu under "The Data Mine" section. You should be presented with a screen similar to figure (3). Select the following settings:
-
Allocation: cis220051
-
Queue: shared
-
Time in Hours: 3
-
Cores: 1
-
Use Jupyter Lab instead of Jupyter Notebook: Checked
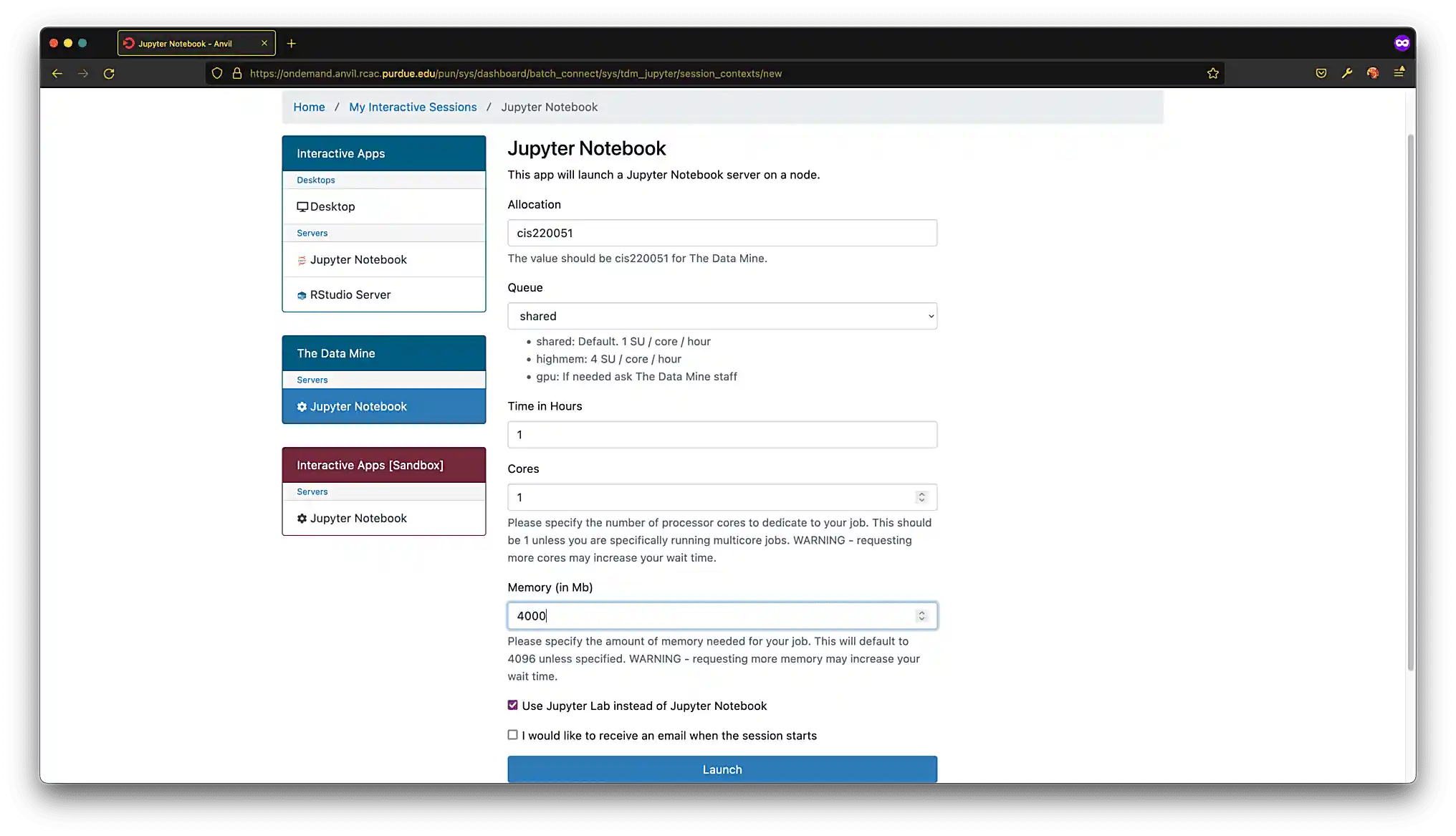 Figure 3. Jupyter Lab settings
Figure 3. Jupyter Lab settings
-
-
When satisfied, click Launch, and wait for a minute. In a few moments, you should be presented with a screen similar to figure (4).
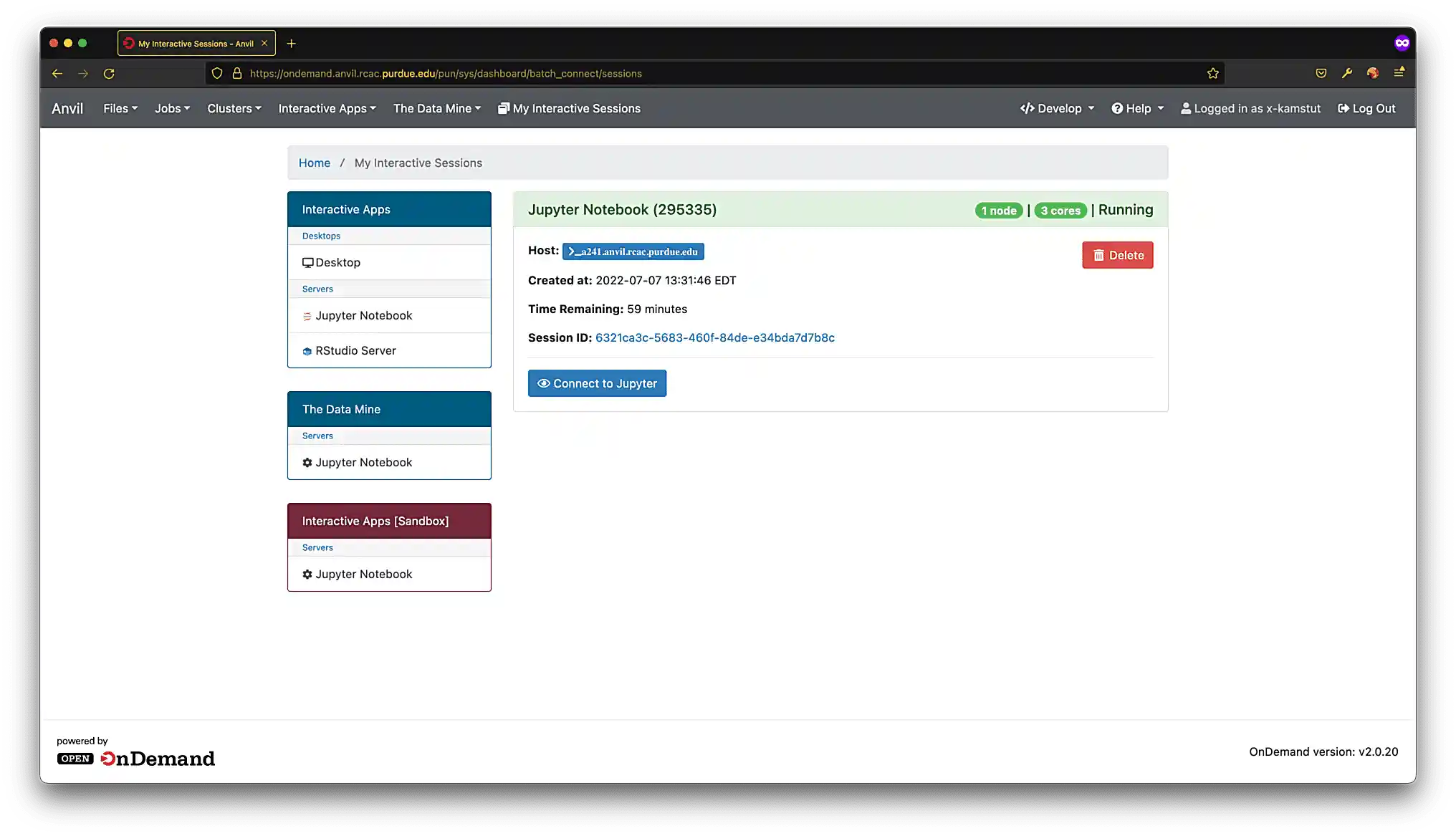 Figure 4. Jupyter Lab ready to connect
Figure 4. Jupyter Lab ready to connect -
When you are ready, click Connect to Jupyter. A new browser tab will launch and you will be presented with a screen similar to figure (5).
 Figure 5. Kernel menu
Figure 5. Kernel menu -
Under the "Notebook" menu, please select the seminar (look for the big "S"; we do not want seminar-r). Finally, you will be presented with a screen similar to figure (6).
 Figure 6. Ready-to-use Jupyter Lab notebook
Figure 6. Ready-to-use Jupyter Lab notebookYou now have a running Jupyter Lab notebook ready for you to use. This Jupyter Lab instance is running on the Anvil cluster. By using OnDemand, you’ve essentially carved out a small portion of the compute power to use. Congratulations! Now please follow along below depending on whether you’d like to do option (1) or option (2).
To run queries directly in a Jupyter Lab cell (1), please do the following.
-
In the first cell, run the following code. This code establishes a connection to the
imdb.dbdatabase, which allows you to directly run SQL queries in a cell as long as that cell has%%sqlat the top of the cell.%sql sqlite:////anvil/projects/tdm/data/movies_and_tv/imdb.db -
After running that cell (for example, using Ctrl+Enter), you can directly run future queries in each cell by starting the cell with
%%sqlin the first line. For example.%%sql SELECT * FROM titles LIMIT 5;While this method has its advantages, there are some advantages to having interop between R and SQL — for example, you could quickly create cool graphics using data in the database and R.
To run queries from within R (2), please do the following.
-
You can directly run R code in any cell that starts with
%%Rin the first line. For example.%%R my_vec <- c(1,2,3) my_vecNow, because we are able to run R code, we can connect to the database, make queries, and build plots, all in a single cell. For example.
%%R library(RSQLite) library(ggplot2) conn <- dbConnect(RSQLite::SQLite(), "/anvil/projects/tdm/data/movies_and_tv/imdb.db") myDF <- dbGetQuery(conn, "SELECT * FROM titles LIMIT 5;") ggplot(myDF) + geom_point(aes(x=primary_title, y=runtime_minutes)) + labs(x = 'Title', y= 'Minutes')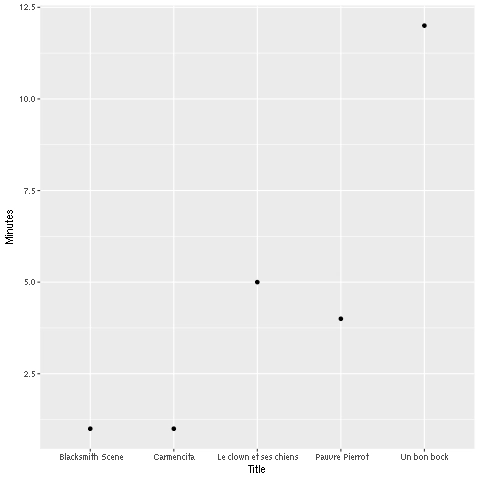 Figure 7. R output
Figure 7. R output
| It is perfectly acceptable to mix and match SQL cells and R cells in your project. |